
Laboratory of Genome Regulation
@Biomedical Pioneering Innovation Center (BIOPIC)
@Beijing Advanced Innovation Center of Genomics (ICG)
@Peking University
Our Research
We aim to learn more about our genome in sequence, conformation, and function.
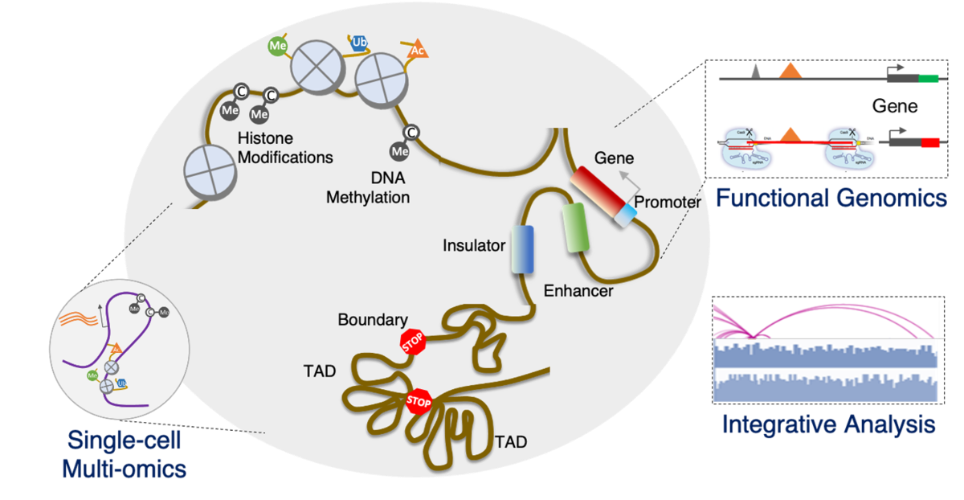
98% of the mammalian genome are non-coding regions, which harbor numerous cis-regulatory elements and facility the spatial organization of the genome. The dynamic of genome organization and cis-regulatory elements contribute to spatiotemporal gene activities that can direct specialized cell types in multi-cellular organisms, which forms the foundation for us to understand the diverse biological processes in different physiological and pathological conditions.
In our lab, we will utilize single-cell multi-omics and functional genomics to examine the mechanisms and functions of epigenetic modifications, 3D genome organization, and non-coding cis-regulatory elements in regulating gene expression, which will be the foundation to further understand gene regulation in cell fate decision, embryo development, and its dysregulation in variety of human diseases.
Representative Publications
For a complete list of publications, click here. (*equal contribution. #co-correspondence)
- Yucen Wang, Zhuoyu Zhang, Guoqiang Li#, Spatial pattern enhanced cellular and tissue recognition for spatial transcriptomics, NAR Genomics and Bioinformatics, 7(2025), lqaf106.
- Hao Tian, Pengfei Luan, Yaping Liu, Guoqiang Li#, Tet-mediated DNA methylation dynamics affect chromosome organization, Nucleic Acids Research, 52(2024): 3654-3666.
- Guoqiang Li*, Yaping Liu*, Yanxiao Zhang, Naoki Kubo, Miao Yu, Rongxin Fang, Manolis Kellis and Bing Ren. “Joint profiling of DNA methylation and chromatin architecture in single cells.” Nature Methods, 16 (2019): 991–993.
- Xiaocui Xu*, Guoqiang Li*, Congru Li*, Jing Zhang*, Qiang Wang*, David K Simmons, Xuepeng Chen, Naveen Wijesena, Wei Zhu, Zhanyang Wang, Zhenhua Wang, Bao Ju, Weimin Ci, Xuemei Lu, Daqi Yu, Qian-fei Wang, Neelakanteswar Aluru, Paola Oliveri, Yong E Zhang, Mark Q Martindale and Jiang Liu. “Evolutionary transition between invertebrates and vertebrates via methylation reprogramming in embryogenesis.” National Science Review, nwz064 (2019)
- Congru Li*, Yang Yu*, Yong Fan*, Guoqiang Li*, Xiaocui Xu, Jialei Duan, Rong Li, Xiangjin Kang, Xin Ma, Xuepeng Chen, Yuwen Ke, Jie Yan, Ying Lian, Ping Liu, Yue Zhao, Hongcui Zhao, Yaoyong Chen, Xiaofang Sun, Jianqiao Liu, Jie Qiao and Jiang Liu. “DNA methylation reprogramming of functional elements during mammalian embryonic development.” Cell Discovery 4, no. 28 (2018).
- Guoqiang Li*, Yang Yu*, Yong Fan*, Congru Li*, Xiaocui Xu, Jialei Duan, Rong Li, Xiangjin Kang, Xin Ma, Xuepeng Chen, Yuwen Ke, Jie Yan, Ying Lian, Ping Liu, Yue Zhao, Hongcui Zhao, Yaoyong Chen, Xiaofang Sun, Jianqiao Liu, Jie Qiao and Jiang Liu. “Genome-wide abnormal DNA methylome of human blastocyst in assisted reproductive technology.” Journal of Genetics and Genomics 44 (2017): 475-481.
- Guoqiang Li*, Weimin Ci*, Subhradip Karmakar*, Ke Chen, Zhixiang Fan, Zhongqiang Guo, Jing Zhang, Lu Wang, Min Zhuang, Shengdi Hu, Xuesong Li, Xianghong Li, Matthew F. Calabrese, Edmond R. Watson, Sandip Prasad, Carrie Rinker-Schaeffer, Scott E. Eggener, Thomas Stricker, Yong Tian, Brenda Schulman, Jiang Liu and Kevin P. White. “SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer.” Cancer Cell 25, no. 4 (2014): 455-468.
Current Members
We’re continuously growing and always looking for motivated students and postdocs to join our team. Please get in touch with the PI if you want to join us.

Huihui Du 杜慧慧
Lab AdministratorM.S. in Management, Renmin University

Yuhe Pei 裴玉禾
Graduate StudentB.S. in Biology, Wuhan University
Zhuoyu Zhang 张卓宇
Graduate StudentB.S. in Biology, Shandong University
Lai Yilin 赖依林
Graduate Student
B.S. in Biology, Peking University
Alumni
Pengfei Luan (栾鹏飞), Visiting Graduate Student, 2021-2025
Hao Tian (田昊), Postdoc, 2021-2024
Wenbo Bai (白文波), Research Assistant, 2021-2023
Yunning Yuan (袁蕴宁), Undergraduate Summer Intern, 2022, 2023
Lin Xi (席琳), Undergraduate Intern, 2023
Contact us
Email:liguoqiang@pku.edu.cn
Phone: 010-62757927
Address: Biomedical Pioneering Innovation Center, Peking University, Beijing, China, 100871
Li-Lab © 2022

















